Projects
Current Projects
PeGaSus
Genomic Assisted breeding for SUStainable agriculture: A benchmark approach. (Proyectos de Generación de Conocmiento: PID2021-123718OB-I00)
There have been many scientific reports evaluating models and accuracy of GAB in plant breeding programs. However, the information on how, when, and why to apply GAB tools in real plant breeding programs is scarce. The main reason for this lack of information is that most plant breeding programs are private, and also GAB applications are in relatively new tools. Here, we aim to apply knowledge driven genomic prediction tools from a empirical public breeding program that will help to deliver general guideline applications of this technology.

GeneBlend
Combining Genomic Approaches to Study Host-Pathogen Relationships in Wheat and Septoria
Spanish Grant CNS2024-154812 (2025-2027) - €144K
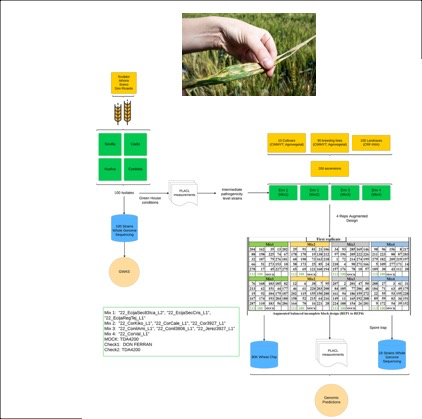
Building on previously developed genomic tools, this project aims to revolutionize our understanding of wheat-Septoria tritici blotch interactions. We're validating predictive models through controlled greenhouse challenges of 200 wheat lines with 100 pathogen isolates, then extending to field conditions with fresh Spanish isolates. Through genome-wide association studies, we're pinpointing resistance loci and developing forecasting tools for pathogen aggressiveness. Our goal is rapid knowledge transfer to breeders and farmers through workshops, training sessions, and publications.
Impact: Enabling breeders to develop durable disease resistance by understanding the genetic architecture of both host and pathogen.
Breed-E-Omics
Genomic Approach for Sustainable Spelt Agriculture (2025-2028). European Grant DADR-2024-029390. (96K).
This project addresses the growing demand for sustainable, high-value spelt wheat. Through comprehensive stakeholder engagement with farmers and agri-food industry, we've identified three critical needs: low-input high-yield varieties for farmers, nutritious organic grains for food processors, and promotion of soil health and biodiversity. We're conducting varietal assessments, genotype-by-environment studies,
and genome-wide association analyses to develop resilient spelt strains that deliver economic, environmental, and social benefits.

OliveMat
Application of Genomic assisted breeding on Olive tree breeding (2025-2029).
In colaboration with University of Cordoba we are aiming to implemente genomic tools to improve genetic
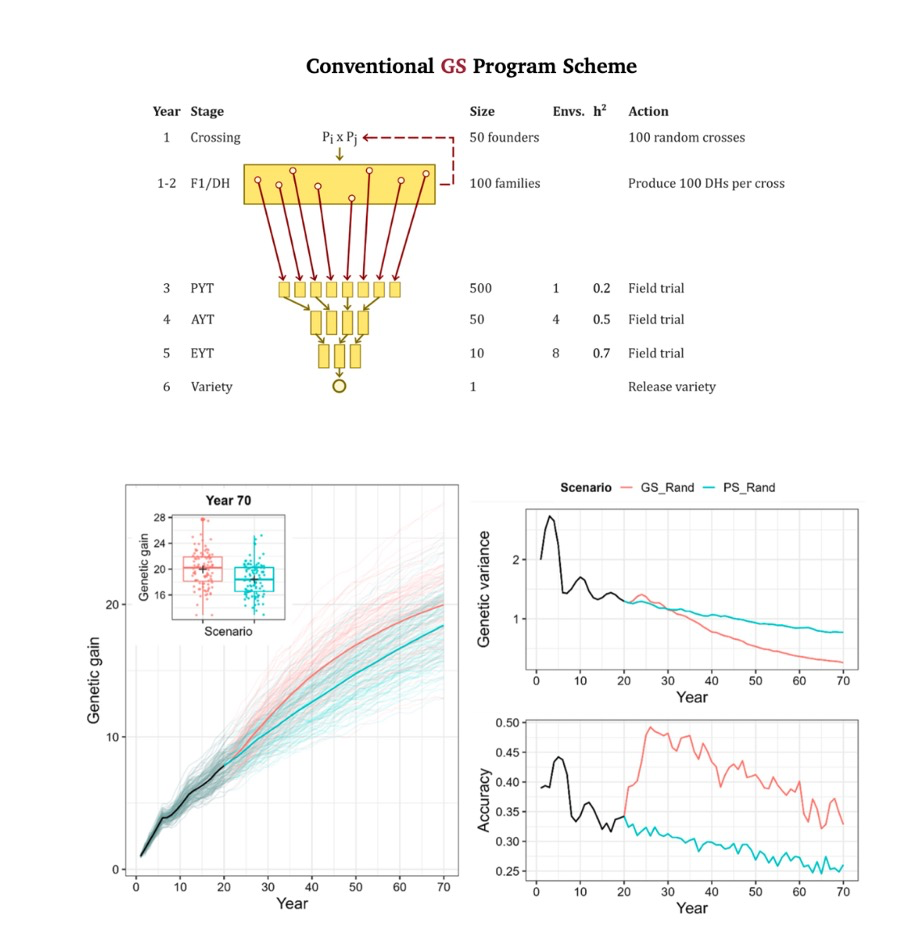
gain on traditional olive tree breeding. A fundamental challenge in breeding is balancing genetic gain against diversity loss. How do we achieve maximum improvement while preserving variation for future progress? Our OGM research leverages stochastic simulations and real-world implementations to support breeders in selecting superior crosses. This approach accelerates development of resilient, high-yielding crops while maintaining long-term genetic diversity—essential for sustained breeding progress.
Paradigm shift: Moving from selecting individual plants to optimizing entire mating strategies for multi-generational impact.
CDTI-Blueberry
Climate-Resilient Blueberry Varieties through Advanced Genomic Selection. Collaboration with Horticulture Company (2023-2026)
This innovative partnership applies cutting-edge genomic selection to fast-track blueberry cultivar development for water stress and warm climates. Over three breeding cycles, we're phenotyping 200-400 seedlings annually for key traits (phenology, vigor, fruit quality) while conducting high-throughput genotyping. Our iteratively refined predictive models enable early identification of superior genotypes, drastically reducing field trial requirements and accelerating breeding cycles.
Innovation: Demonstrating genomic selection's power beyond traditional field crops—expanding to high-value horticultural species.

Machine learning approaches applied to genomic assisted breeding
Programa Propio de I+D+I 20201 de la Universidad Politécnica de Madrid.
Convocatoria de ayudas para contratos predoctorales.
Wheat (the second most important crop worldwide) yields are not currently increasing at comparable rates to those achieved in previous decades. Exploitation of the full range of available genetic resources (pre-breeding) could help develop new varies that will be needed in the future. The emergent technologies based on Artificial Intelligence and Machine learning are interesting tools to handle the huge volumes of genotypic and phenotypic data that are generated from agronomic and biological experiments. The aim of this PhD project is to develop statistical and programming tools to exploit and decode the hidden patterns underlying data and apply this knowledge on real breeding problems in both academical and industrial fields.

Genomic assisted breeding applied to Syngenta sunflower breeding program.
Programa UPM-Syngenta.
Convocatorias FPU
This project aims to provide a benchmark guideline on genomic assisted breeding for Syngenta sunflowerbreeding program by building tools and models that augment current practices capitalizing on advances in genomics, phenomics, imaging technologies, and machine learning.

Previous Projects
European Grants
INNOVAR: Next Generation Variety Testing For Improved Cropping On European Farmland (H2020) (2020-2025)
Granted: 350 K. PI. Julio Isidro-Sánchez
HealthyOats: Ireland Wales 2014-2020 Operation
Granted: 2 million. Coordinator. Julio Isidro-Sánchez
WheatSustain: Knowledge-driven genomic predictions for sustainable disease resistance in wheat (Suscrop)
Granted: 173K. PI. Julio Isidro-Sánchez
Identification and selection of traits that maximize biomass production as well as
enhance the efficiency of biomass conversion by novel processes are critical for the
viability of biorefineries (2009-2011)
AGRNEX2008N0475. Postdoctoral Project.
IDuWUE: Improving durum wheat for water use efficiency and yield stability through physiological and molecular approaches (2002-2006)
ICFPN502A3PR03 (ICA3NCTN2002N10085), 963K. PhD. Project.
Irish Grants
Oats for the future: Deciphering potential of host resistance and RNAi to minimise mycotoxin contamination under present and future climate scenarios
Granted: 89K. PI. Julio Isidro-Sánchez
Developing multi-use barley to improve the organic Irish market (2019-2022)
Granted-96K. PI. Julio Isidro-Sánchez.
PICS: Physiology Infrastructure for Crop Stress (2016)
295K. PI. Julio Isidro-Sánchez.
Canadian Grants
CTAG: Canadian Triticum Applied Genomics (2015-2019)
Scientific advisor of WPs.
Improving Wheat Productivity under Conditions of Abiotic Stress (2012-2017)
A proposed
project as part of the National Research Council Wheat Flagship Program and the Canadian Wheat
Improvement Consortium. 14 million. Postdoctoral Project.
Spanish Grants
WheatRes. Identificación de nuevas fuentes de resistencia horizontal a septoria y roya en trigo duro (2021-2024)
PLEC2021-007930. 160K. PI on this project.
Aproximación multidisciplinary al incremento de la eficacia en la mejora del trigo
duro: Integración de tácnicas ecofisiológicas y moleculares (2003-2006)
AGL2002-04285-C03-02. 65K. Member of the Research team of this project.
Nuevas vias para mejorar la adaptacion del trigo duro, Triticum turgidum L.Var.Durum
a ambientes mediterraneos (2006-2009)
AGL2006-09226-C02-02- 02/AGR. 45K. Member of the Research team of this project.
Fisiología del tritórdeo en condiciones mediterráneas: enfoque multidisciplinary
para una colaboración transmediterránea azahar (2005-2008)
AGL2005-07257- C04-04. 28K. Member of the Research team of this project.

























































